* force add PATH to current user * checkin docker setup script * Update cluster_setup.sh * install docker and start container on cluster setup * WIP: Run task in container * fix merge conflict * run tasks and merge task from within container * refactor code to proper docker commands and make a single R container per job * refactor command line utils into its own file * refactor job utilities into its own file * move cluster setup script to inst folder * remove unnecessary curl installs * remove starting container from setup script * check in WIP * add apt_install file * make required directories * update cluster setup files as needed * include libxml2 packages in apt installs * working cluster create with cran and github dependencies * update job prep to install apt-get and not each task * use rocker containers instead of r-base * remove unused & commented code * remove unused install function * address several lintr issues * initial test dockerfile * add spacing between commands * temporarily point wget to feature branch * update bioconductor install for non-jobPrep installs * Delete Dockerfile * minor changes to install bioc * resolve merge conflicts * update cluster to correctly install BioC packages using install_bioconductor * fix issue where some packages were not getting installed * add missing BioConductorCommand initializer * remove print lines * initial dockerfile implementations * update docker files * Only install packages if they are required * Remove requirement on bioconductor installer script on start task * remove duplicate environment variable entry * update docs for container support * update version to 0.6.0 * refactor changes updates * remove poorly formatted whitespaces * add full path to pacakges directory * fix docker command line * update file share sample * update azure files cluster name * update mandelbrot sample * update package management sample * update plyr samples * make montecarlo sample more consistent * update montecarlo sample * remove plyr example * fix bad environment pointer * fix linter issues * more linter fixes * more linter issues * use latest rAzureBatch version * update resource files example * remove reference to deleted sample * pr feedback * PR docs feedback * Print errors from worker (#154) * Fixed pool package command line lintr test * Package installation tests fixed - too long lines * Fixed json in customize cluster docs * Fix: Typos in customize cluster docs * Cleaning up files * Feature/githubbiopackage (#150) * install github package worked for foreach loop * fix lintr error * tests for github and bioc packages installation * lintr fix * add back lost code due to merge and update docs * The Travis CI build failed for feature/githubbiopackage * remove incorrect parameter for install_github * Updated job prep task to have default command * Use the latest version of rAzureBatch * Updated description + Generate cluster config * Fix: Bioconductor and Github packages installation (#155) * Added multiple package install test and fix obj reading args * Fixed naming for packages install * Replaced validation exclusion for linter * Fixed test validate test * Fixing all interactive tests with skip * Fixed renaming validation * Removed default test - cannot be tested * Removed in validation * Added cluster package install tests (#156) |
||
|---|---|---|
| R | ||
| docker-image | ||
| docs | ||
| inst/startup | ||
| man | ||
| samples | ||
| tests | ||
| vignettes | ||
| .Rbuildignore | ||
| .gitattributes | ||
| .gitignore | ||
| .lintr | ||
| .travis.yml | ||
| CHANGELOG.md | ||
| Contributing.md | ||
| DESCRIPTION | ||
| LICENSE | ||
| NAMESPACE | ||
| README.md | ||
README.md
doAzureParallel
# set your credentials
setCredentials("credentials.json")
# setup your cluster with a simple config file
cluster<- makeCluster("cluster.json")
# register the cluster as your parallel backend
registerDoAzureParallel(cluster)
# run your foreach loop on a distributed cluster in Azure
number_of_iterations <- 10
results <- foreach(i = 1:number_of_iterations) %dopar% {
myParallelAlgorithm()
}
Introduction
The doAzureParallel package is a parallel backend for the widely popular foreach package. With doAzureParallel, each iteration of the foreach loop runs in parallel on an Azure Virtual Machine (VM), allowing users to scale up their R jobs to tens or hundreds of machines.
doAzureParallel is built to support the foreach parallel computing package. The foreach package supports parallel execution - it can execute multiple processes across some parallel backend. With just a few lines of code, the doAzureParallel package helps create a cluster in Azure, register it as a parallel backend, and seamlessly connects to the foreach package.
NOTE: The terms pool and cluster are used interchangably throughout this document.
Dependencies
- R (>= 3.3.1)
- httr (>= 1.2.1)
- rjson (>= 0.2.15)
- RCurl (>= 1.95-4.8)
- digest (>= 0.6.9)
- foreach (>= 1.4.3)
- iterators (>= 1.0.8)
- bitops (>= 1.0.5)
Installation
Install doAzureParallel directly from Github.
# install the package devtools
install.packages("devtools")
# install the doAzureParallel and rAzureBatch package
devtools::install_github("Azure/doAzureParallel")
Azure Requirements
To run your R code across a cluster in Azure, we'll need to get keys and account information.
Setup Azure Account
First, set up your Azure Account (Get started for free!)
Once you have an Azure account, you'll need to create the following two services in the Azure portal:
- Azure Batch Account (Create an Azure Batch Account in the Portal)
- Azure Storage Account (this can be created with the Batch Account)
Get Keys and Account Information
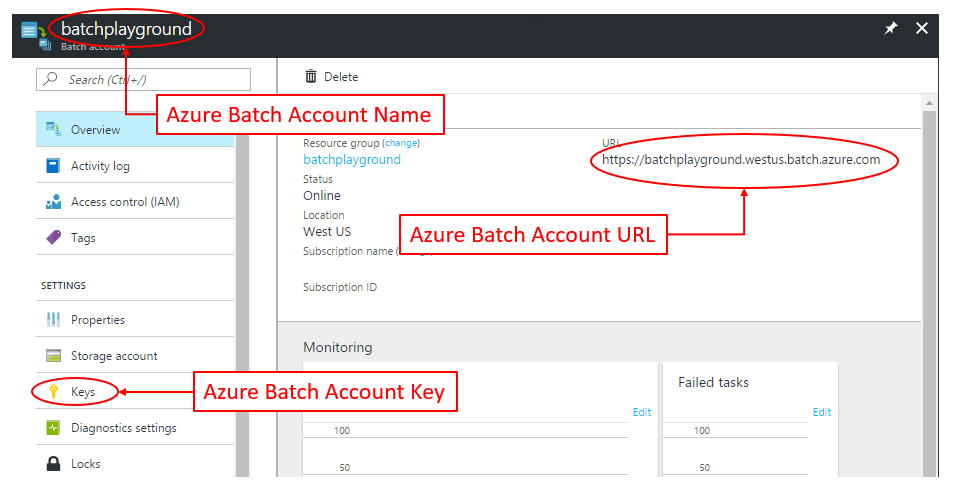
For your Azure Batch Account, we need to get:
- Batch Account Name
- Batch Account URL
- Batch Account Access Key
This information can be found in the Azure Portal inside your Batch Account:
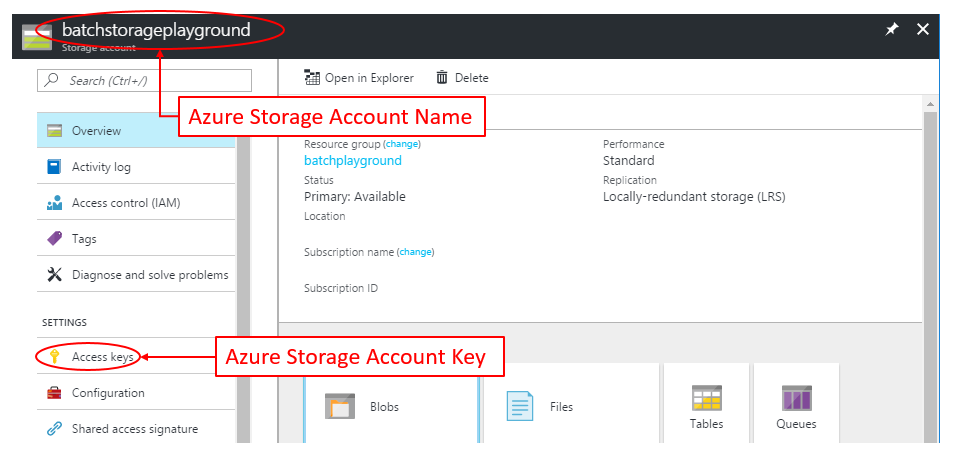
For your Azure Storage Account, we need to get:
- Storage Account Name
- Storage Account Access Key
This information can be found in the Azure Portal inside your Azure Storage Account:
Keep track of the above keys and account information as it will be used to connect your R session with Azure.
Getting Started
Import the package
library(doAzureParallel)
Set up your parallel backend with Azure. This is your set of Azure VMs.
# 1. Generate your credential and cluster configuration files.
generateClusterConfig("cluster.json")
generateCredentialsConfig("credentials.json")
# 2. Fill out your credential config and cluster config files.
# Enter your Azure Batch Account & Azure Storage keys/account-info into your credential config ("credentials.json") and configure your cluster in your cluster config ("cluster.json")
# 3. Set your credentials - you need to give the R session your credentials to interact with Azure
setCredentials("credentials.json")
# 4. Register the pool. This will create a new pool if your pool hasn't already been provisioned.
cluster <- makeCluster("cluster.json")
# 5. Register the pool as your parallel backend
registerDoAzureParallel(cluster)
# 6. Check that your parallel backend has been registered
getDoParWorkers()
Run your parallel foreach loop with the %dopar% keyword. The foreach function will return the results of your parallel code.
number_of_iterations <- 10
results <- foreach(i = 1:number_of_iterations) %dopar% {
# This code is executed, in parallel, across your cluster.
myAlgorithm()
}
After you finish running your R code in Azure, you may want to shut down your cluster of VMs to make sure that you are not being charged anymore.
# shut down your pool
stopCluster(cluster)
Configuration JSON files
Credentials
Use your credential config JSON file to enter your credentials.
{
"batchAccount": {
"name": <Azure Batch Account Name>,
"key": <Azure Batch Account Key>,
"url": <Azure Batch Account URL>
},
"storageAccount": {
"name": <Azure Storage Account Name>,
"key": <Azure Storage Account Key>
}
}
Learn more:
Cluster Settings
Use your pool configuration JSON file to define your pool in Azure.
{
"name": <your pool name>, // example: "myazurecluster"
"vmSize": <your pool VM size name>, // example: "Standard_F2"
"maxTasksPerNode": <num tasks to allocate to each node>, // example: "2"
"poolSize": {
"dedicatedNodes": { // dedicated vms
"min": 2,
"max": 2
},
"lowPriorityNodes": { // low priority vms
"min": 1,
"max": 10
},
"autoscaleFormula": "QUEUE"
},
"rPackages": {
"cran": ["some_cran_package", "some_other_cran_package"],
"github": ["username/some_github_package", "another_username/some_other_github_package"],
"githubAuthenticationToken": {}
},
"commandLine": []
}
NOTE: If you do not want your cluster to autoscale, simply set the number of min nodes equal to max nodes for low-priority and dedicated.
Learn more:
Low Priority VMs
Low-priority VMs are a way to obtain and consume Azure compute at a much lower price using Azure Batch. Since doAzureParallel is built on top of Azure Batch, this package is able to take advantage of low-priority VMs and allocate compute resources from Azure's surplus capacity at up to 80% discount.
Low-priority VMs come with the understanding that when you request it, there is the possibility that we'll need to take some or all of it back. Hence the name low-priority - VMs may not be allocated or may be preempted due to higher priority allocations, which equate to full-priced VMs that have an SLA.
And as the name suggests, this significant cost reduction is ideal for low priority workloads that do not have a strict performance requirement.
With Azure Batch's first-class support for low-priority VMs, you can use them in conjunction with normal on-demand VMs (dedicated VMs) and enable job cost to be balanced with job execution flexibility:
- Batch pools can contain both on-demand nodes and low-priority nodes. The two types can be independently scaled, either explicitly with the resize operation or automatically using auto-scale. Different configurations can be used, such as maximizing cost savings by always using low-priority nodes or spinning up on-demand nodes at full price, to maintain capacity by replacing any preempted low-priority nodes.
- If any low-priority nodes are preempted, then Batch will automatically attempt to replace the lost capacity, continually seeking to maintain the target amount of low-priority capacity in the pool.
- If tasks are interrupted when the node on which it is running is preempted, then the tasks are automatically re-queued to be re-run.
For more information about low-priority VMs, please visit the documentation.
You can also check out information on low-priority pricing here.
Distributing Data
When developing at scale, you may also want to chunk up your data and distribute the data across your nodes. Learn more about that here
Using %do% vs %dopar%
When developing at scale, it is always recommended that you test and debug your code locally first. Switch between %dopar% and %do% to toggle between running in parallel on Azure and running in sequence on your local machine.
# run your code sequentially on your local machine
results <- foreach(i = 1:number_of_iterations) %do% { ... }
# use the doAzureParallel backend to run your code in parallel across your Azure cluster
results <- foreach(i = 1:number_of_iterations) %dopar% { ... }
Error Handling
The errorhandling option specifies how failed tasks should be evaluated. By default, the error handling is 'stop' to ensure users' can have reproducible results. If a combine function is assigned, it must be able to handle error objects.
| Error Handling Type | Description |
|---|---|
| stop | The execution of the foreach will stop if an error occurs |
| pass | The error object of the task is included the results |
| remove | The result of a failed task will not be returned |
# Remove R error objects from the results
res <- foreach::foreach(i = 1:4, .errorhandling = "remove") %dopar% {
if (i == 2 || i == 4) {
randomObject
}
mean(1:3)
}
#> res
#[[1]]
#[1] 2
#
#[[2]]
#[1] 2
# Passing R error objects into the results
res <- foreach::foreach(i = 1:4, .errorhandling = "pass") %dopar% {
if (i == 2|| i == 4) {
randomObject
}
sum(i, 1)
}
#> res
#[[1]]
#[1] 2
#
#[[2]]
#<simpleError in eval(expr, envir, enclos): object 'randomObject' not found>
#
#[[3]]
#[1] 4
#
#[[4]]
#<simpleError in eval(expr, envir, enclos): object 'randomObject' not found>
Long-running Jobs + Job Management
doAzureParallel also helps you manage your jobs so that you can run many jobs at once while managing it through a few simple methods.
# List your jobs:
getJobList()
# Get your job by job id:
getJob(jobId = 'unique_job_id', verbose = TRUE)
This will also let you run long running jobs easily.
With long running jobs, you will need to keep track of your jobs as well as set your job to a non-blocking state. You can do this with the .options.azure options:
# set the .options.azure option in the foreach loop
opt <- list(job = 'unique_job_id', wait = FALSE)
# NOTE - if the option wait = FALSE, foreach will return your unique job id
job_id <- foreach(i = 1:number_of_iterations, .options.azure = opt) %dopar % { ... }
# get back your job results with your unique job id
results <- getJobResult(job_id)
Finally, you may also want to track the status of jobs by state (active, completed etc):
# List jobs in completed state:
filter <- list()
filter$state <- c("active", "completed")
jobList <- getJobList(filter)
View(jobList)
You can learn more about how to execute long-running jobs here.
With long-running jobs, you can take advantage of Azure's autoscaling capabilities to save time and/or money. Learn more about autoscale here.
Using the 'chunkSize' option
doAzureParallel also supports custom chunk sizes. This option allows you to group iterations of the foreach loop together and execute them in a single R session.
# set the chunkSize option
opt <- list(chunkSize = 3)
results <- foreach(i = 1:number_of_iterations, .options.azure = opt) %dopar% { ... }
You should consider using the chunkSize if each iteration in the loop executes very quickly.
If you have a static cluster and want to have a single chunk for each worker, you can compute the chunkSize as follows:
# compute the chunk size
cs <- ceiling(number_of_iterations / getDoParWorkers())
# run the foreach loop with chunkSize optimized
opt <- list(chunkSize = cs)
results <- foreach(i = 1:number_of_iterations, .options.azure = opt) %dopar% { ... }
Resizing Your Cluster
At some point, you may also want to resize your cluster manually. You can do this simply with the command resizeCluster.
cluster <- makeCluster("cluster.json")
# resize so that we have a min of 10 dedicated nodes and a max of 20 dedicated nodes
# AND a min of 10 low priority nodes and a max of 20 low priority nodes
resizeCluster(
cluster,
dedicatedMin = 10,
dedicatedMax = 20,
lowPriorityMin = 10,
lowPriorityMax = 20,
algorithm = 'QUEUE',
timeInterval = '5m' )
If your cluster is using autoscale but you want to set it to a static size of 10, you can also use this method:
# resize to a static cluster of 10
resizeCluster(cluster,
dedicatedMin = 10,
dedicatedMax = 10,
lowPriorityMin = 0,
lowPriorityMax = 0)
Setting Verbose Mode to Debug
To debug your doAzureParallel jobs, you can set the package to operate on verbose mode:
# turn on verbose mode
setVerbose(TRUE)
# turn off verbose mode
setVerbose(FALSE)
Bypassing merge task
Skipping the merge task is useful when the tasks results don't need to be merged into a list. To bypass the merge task, you can pass the enableMerge flag to the foreach object:
# Enable merge task
foreach(i = 1:3, .options.azure = list(enableMerge = TRUE))
# Disable merge task
foreach(i = 1:3, .options.azure = list(enableMerge = FALSE))
Note: User defined functions for the merge task is on our list of features that we are planning on doing.
Next Steps
For more information, please visit our documentation.